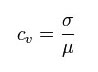ELISA Test
As to better illustrate how ELISA test works, this section took a deeper insight into: a definition of the 'sensitivity', standard curve calculation, and control samples in ELISA test, followed by a list of diseases in which ELISA test could be applied.
ELISA Test-Sensitivity
The factors that determine the ultimate sensitivity of a competitive ELISA test are the antibody affinity constant and the experimental errors. The detection limit of the substrate is not typically limiting. It has been calculated theoretically that with a K = 1012 M-1 (an extraordinarily high equilibrium constant for an antigen-antibody interaction) and a 1% co-efficient of variation (CV) for the response at zero dose, the lowest detection limit possible would be 10-14 M. A more easily achievable limit would be 10-9 - 10-10 M.
The factors limiting the sensitivity of a sandwich ELISA test are the affinity of the antibody, the experimental errors and the nonspecific binding of the labeled antibody, expressed as a percentage of the total antibody. It has been estimated that with a K = 1012 M-1, 1% CV of the response at zero dose, and a 1% nonspecific binding of the labeled antibody, the detection limit can be as low as 10-16 M. In addition, this can be enhanced further by using more sensitive detection substrates.

This is expressed as a percentage of variance to the mean and therefore indicates any inconsistencies and inaccuracies in the results. This sets a standard for the quality of the validated results. Computer programs can be used to calculate the CV values from ELISA test results.
ELISA Test-Control samples
• Positive control
As for the positive control in an ELISA test, use either an endogenous soluble sample known to contain the protein you are detecting, or a purified protein or peptide known to contain the immunogen sequence for the antibody you are using. A positive result from the positive control, even if the samples are negative, will indicate the procedure is optimized and working. It will verify that any negative results are valid. Any tissues, cells or lysates that have been used successfully can be considered a suitable positive control. Try looking at the Swiss-Prot or Omnigene database links on the datasheet. These databases will often have a list of tissues that the protein is expressed in. These can also be considered suitable positive controls. Check the GeneCards entry for the protein. This will usually provide you with relative levels of expression in various tissues. If you still have difficulty finding a suitable control, we recommend doing a quick literature search on PubMed to see which tissues and cells express the protein of interest.
• Negative control
The negative control in an ELISA test means a sample known not to express the protein you are detecting. This is to check for non-specific binding and false positive results. Each plate you use should contain a negative control sample in order to validate the results.
• Standard
A sample containing a known concentration of the target protein from which the standard curve can be obtained.
• Standard in sample matrix (spike control)
When testing serum samples in ELISA tests, include a standard in normal diluent buffer as usual. But we recommend to also include a standard diluted in serum from the species you are testing. The two can then be compared to ensure there is no effect on the standard curve from other proteins in the serum. This is known as a spike control.
• Endogenous positive control
We recommend including an endogenous positive control if you are testing a recombinant protein sample. This should be an essential component of your experiment.
There are inherent difficulties with antibody detection of recombinant proteins that need to be considered. Folding of the recombinant protein may be different from the endogenous native form, and may prevent antibody access to the epitope. This is particularly the case with tagged proteins. Always ensure tags are placed on the N or C terminal end of the recombinant protein.
Most importantly, always ensure the recombinant protein includes the immunogen sequence of the antibody you are using. An endogenous positive control is important to validate the results, as well as to indicate how well the reagents (eg antibodies) and procedure are working.